research use only
LDN-57444 DUB inhibitor
Cat.No.S7135
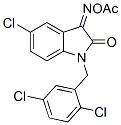
Chemical Structure
Molecular Weight: 397.64
Quality Control
| Related Targets | HDAC Caspase Proteasome Secretase MMP HCV Protease Cysteine Protease DPP Tyrosinase HIV Protease |
|---|---|
| Other DUB Inhibitors | PR-619 P5091 IU1 P22077 b-AP15 ML323 VLX1570 TCID EOAI3402143 ML364 |
Chemical Information, Storage & Stability
| Molecular Weight | 397.64 | Formula | C17H11Cl3N2O3 |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 668467-91-2 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | N/A | Smiles | CC(=O)ON=C1C2=C(C=CC(=C2)Cl)N(C1=O)CC3=C(C=CC(=C3)Cl)Cl | ||
Solubility
|
In vitro |
DMSO
: 11 mg/mL
(27.66 mM)
Water : Insoluble Ethanol : Insoluble |
Molarity Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such as vortex, ultrasound or hot water bath can be used to aid dissolving.
Mechanism of Action
| Targets/IC50/Ki |
UCH-L1
0.4 μM(Ki)
UCH-L1
0.88 μM
UCH-L3
25 μM
|
|---|---|
| In vitro |
Treatment with 50 μM LDN-57444 for 24 h leads to 70% inhibition of the proteasome activity. This compound causes a significant and concentration-dependent decrease in cell viability at concentrations above 25 μM and the cell viability reduced to 61.81% at 50 μM. It is able to cause cell death through the apoptosis pathway by decreasing the activity of ubiquitin proteasome system and increasing the levels of highly ubiquitinated proteins, both of which can activate unfolded protein response. The apoptosis induced by this chemical may be triggered by the activation of endoplasmic reticulum stress (ERS).
|
| Kinase Assay |
HTS screen
|
|
To start an assay, 0.5 μL of 5 mg/mL test compound (about 50 μM final reaction concentration) or DMSO control is aliquoted into each well. Both enzyme and substrate are prepared in UCH reaction buffer (50 mM Tris-HCl [pH 7.6], 0.5 mM EDTA, 5 mM DTT, and 0.5 mg/mL ovalbumin). 25 μL of 0.6 nM UCH-L1 is then added to each well except substrate control wells, followed by plate shaking for 45–60 s on an automatic shaker. The enzyme/compound mixture is incubated at room temperature for 30 min before 25 μL of 200 nM Ub-AMC is added to initiate the enzyme reaction. The reaction mixture (300 pM UCH-L1, 100 nM Ubiquitin-AMC with 2.5 μg test compound) is incubated at room temperature for 30 additional minutes prior to quenching the reaction by the addition of 10 μL 500 mM acetic acid per well. The fluorescence emission intensity is measured on a LJL Analyst using a coumarin filter set (ex = 365 nm, em = 450 nm) and is subtracted by the intrinsic compound fluorescence to reveal the enzyme activity. A DMSO control (0.5 μL of DMSO, 25 μL of UCH-L1, 25 μL of ubiquitin-AMC, 10 μL of acetic acid), enzyme control (25 μL of UCH-L1, 25 μL of buffer, 10 μL of acetic acid), substrate control (25 μL of buffer, 25 μL of ubiquitin-AMC, 10 μL of acetic acid), and inhibitor control (0.5 μL of ubiquitin aldehyde [100 nM stock], 25 μL of UCH-L1, 25 μL of ubiquitin-AMC, 10 μL of acetic acid) are also performed in each assay plate to ensure quality and reproducibility. Potential UCH-L1 inhibitors are selected if the compounds demonstrated greater than 60% inhibition compared to the controls. The UCH-L1 enzymatic reactions are manually repeated twice using the same protocol to confirm the results for the hit compounds from the primary robot-assisted screen.
|
|
| In vivo |
LDN-57444 causes dramatic alterations in synaptic protein distribution and spine morphology in vivo. Treatment with this compound also results in a rapid fall of Uch-L1 activity, but proteasome inhibition has no effect on cAMP levels over a period of several hours.
|
References |
|
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.
